Target Deconvolution Through Data Integration: Unifying Drug and Genetic Perturbations
Tahoe-100M links drug and genetic perturbation maps to rapidly deconvolute targets, expose hidden pathways, and guide design of drugs that phenocopy desired cellular states.
For decades, researchers have struggled with the fact that drugs rarely act on a single target. Polypharmacology defines how drugs really behave in patients, yet systematically mapping those effects has been slow and incomplete. Landmark resources like the Connectivity Map demonstrated that transcriptional profiling could connect drugs to mechanisms, but these efforts were limited in scale and largely based on bulk measurements.
Tahoe 100M changes the equation. This atlas offers millions of transcriptional profiles at single-cell resolution across thousands of compounds, capturing drug effects with unprecedented depth. When combined with CRISPR-based CROP-seq, drug and genetic perturbations can finally be compared head-to-head — creating a unified map for target deconvolution, pathway mapping, and phenotypic drug design.
We combined:
Tahoe-100M drug atlas, a massive single-cell reference of ~400 compounds tested across 50 cancer models, each at three doses. This scale captures a spectrum of dose-dependent drug responses and provides a uniquely rich landscape of transcriptional perturbations.
Myllia’s CROP-seq dataset, a focused CRISPR perturbation dataset in which 218 genes were targeted in A549 cells and transcriptomic fingerprints were recorded by single-cell RNA sequencing.
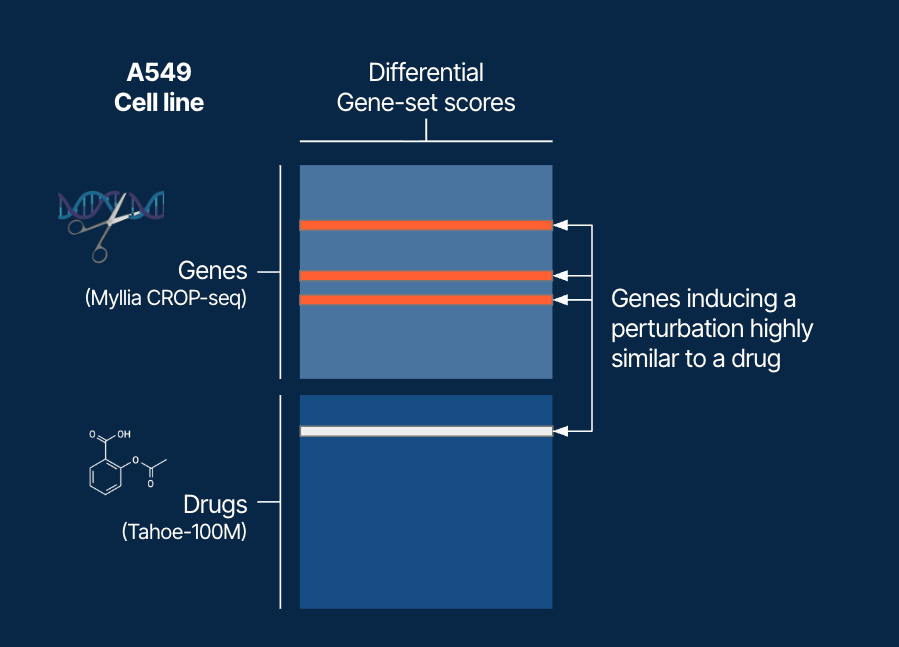
Both were converted into gene-set scores (MSigDB collections, scored with Vision), reducing dimensionality and pooling genes into functional units. Differential scores were computed by comparing drug treatment to matched DMSO (Tahoe-100M), or gene perturbations to non-targeting guides (Myllia’s CROP-seq). Pairwise similarities between drug-induced and gene-induced differential signatures produced a drug–gene similarity matrix (Figure 1).
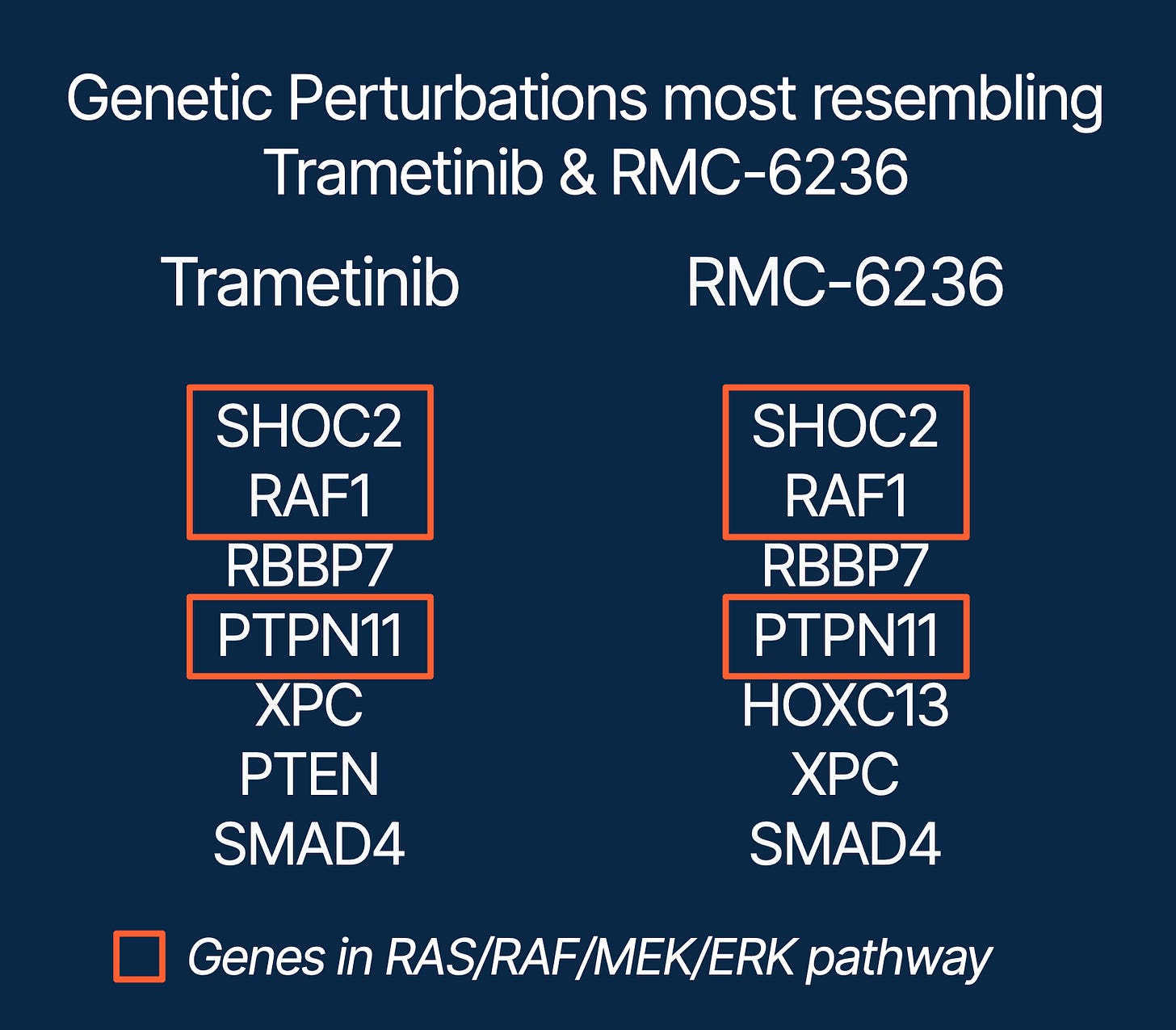
As a first test, we applied the framework to two well-characterized inhibitors:
Trametinib (MEK1/2 inhibitor)
RMC-6236 (pan-RAS inhibitor)
Both landed where they should: the top-ranked genetic perturbations mapped to the RAS/RAF/MEK/ERK pathway, consistent with their known mechanisms of action (Figure 2).
Importantly, MEK1/2 and KRAS themselves were not included in the CRISPR knockout set. Yet, despite their absence, the framework still identified the correct pathway-level convergence through related nodes in the cascade – demonstrating that the integration can resolve mechanisms even when direct targets aren’t explicitly perturbed.
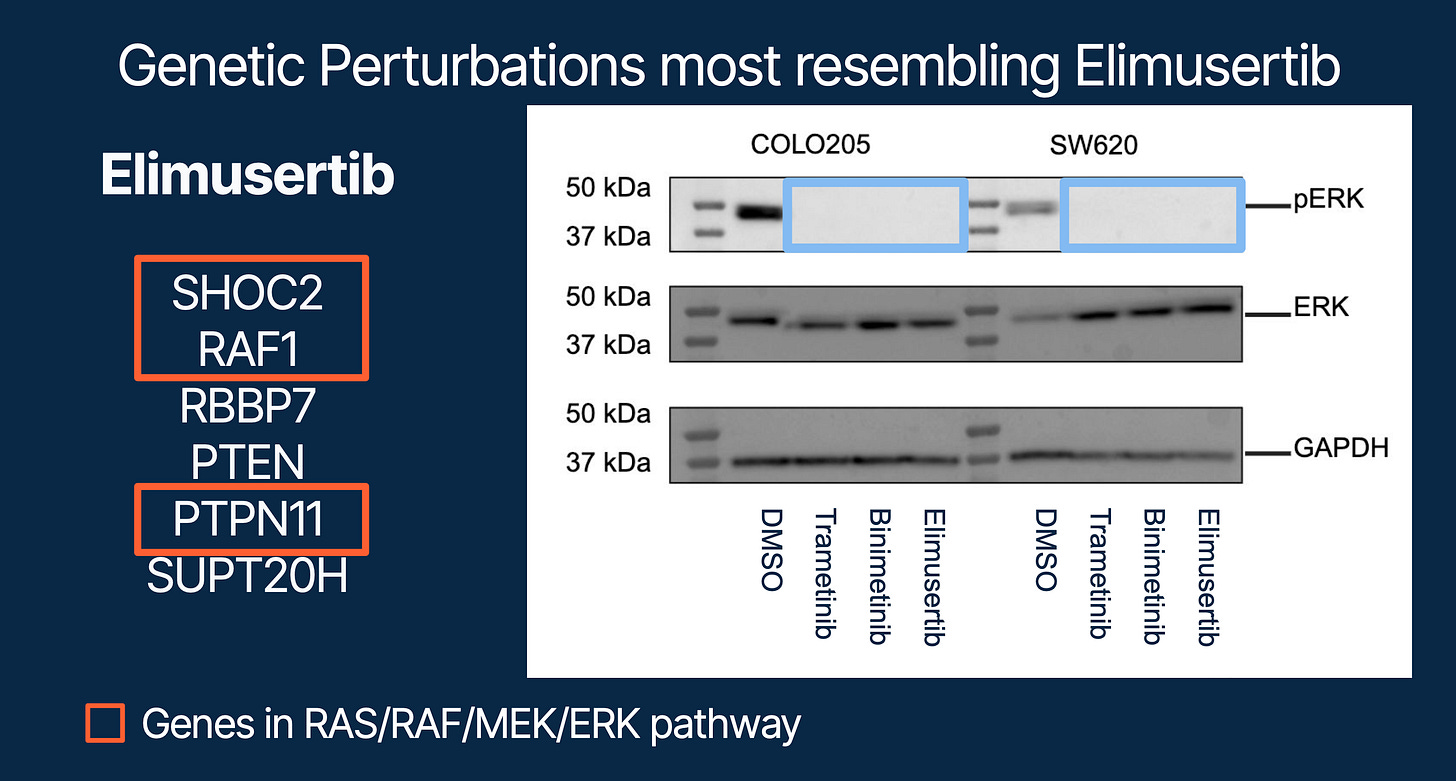
Then came the surprise. Elimusertib, long reported as an ATR inhibitor, did not resemble DNA damage pathway perturbations at all. Instead, its transcriptional profile clustered tightly with MEK inhibitors (Figure 3).
To test this, we treated two cancer cell lines with Elimusertib and measured ERK phosphorylation (pERK). The result: Elimusertib suppressed pERK, phenocopying Trametinib and Binimetinib.
This was a striking twist. A compound presumed to act via ATR was revealed to moonlight as a MAPK pathway suppressor.
And it raises a broader question: what other compounds hide secondary mechanisms, silently shaping efficacy or toxicity? Traditionally, testing every possible mechanism is infeasible; the space is too vast. This integrative approach makes it possible to scan hundreds of pathways at once, rapidly surfacing the most likely hidden effects.
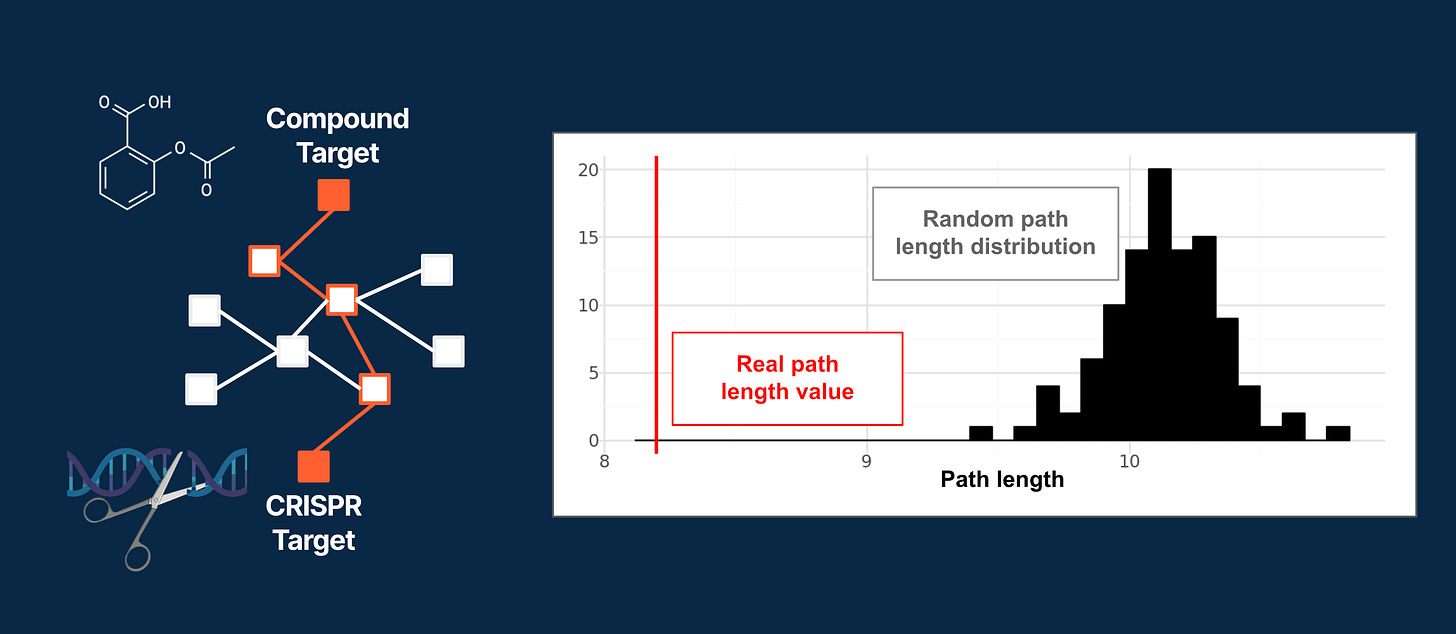
To generalize further, we assessed whether drugs and CRISPR perturbations with functionally related targets tend to elicit similar transcriptional effects. Using the HumanNetV3 functional gene network, we calculated the shortest paths from CRISPR-perturbed genes to drugs producing a similar gene expression response.
The average observed path length was consistently much shorter than expected by chance (compared to random drug sampling), showing that transcriptional similarity reflects true functional proximity (Figure 4).
This work demonstrates a new way of seeing drug action:
Target deconvolution – drug effects can be mapped to specific pathways by reference to genetic perturbations.
Unmasking moonlighting compounds – Elimusertib shows how unexpected mechanisms surface rapidly.
Network convergence – transcriptional similarities aren’t random; they reflect underlying biological relationships.
Phenocopy design – the next frontier is designing compounds that deliberately recreate specific transcriptional states.
Taken together, these findings suggest that the integration of drug and genetic perturbations at scale is more than a technical advance. It’s a conceptual shift: from thinking of drugs as single-target agents, to thinking of them as programmable phenotypic levers capable of reproducing desired cellular states.