Using Tahoe-100M to find new immunomodulators
How the world’s largest single-cell transcriptomic dataset unlocked drugs that enhance tumor visibility through MHC-I upregulation.
Cancer cells evade immune detection by downregulating MHC-I expression to escape immune surveillance. MHC-I molecules are pivotal in presenting tumor-associated antigens to CD8+ cytotoxic T-cells, marking tumor cells for destruction (Figure 1). That’s why MHC-I downregulation poses a critical challenge for immunotherapy.
Successfully overcoming this evasion mechanism could revolutionize cancer treatment by significantly enhancing the effectiveness of immune checkpoint inhibitors (ICIs), which depend on robust tumor antigen presentation. Addressing this fundamental challenge—identifying compounds that effectively upregulate MHC-I expression—can dramatically enhance therapeutic strategies.
Figure 1. Schematic illustrating MHC-I upregulation and effects on immune cell engagement.
We took a crack at that by digging into our Tahoe-100M, the world's largest drug-perturbed, single-cell transcriptomic dataset.
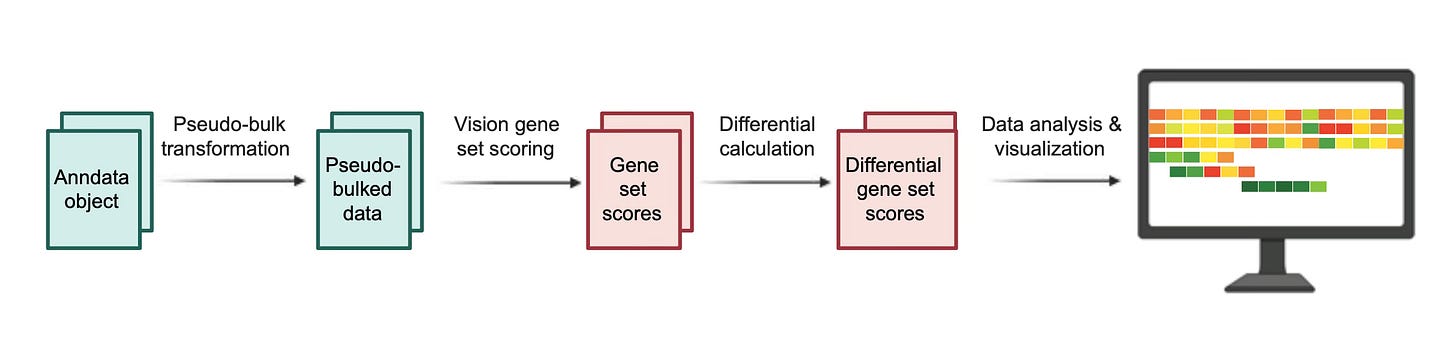
With an unprecedented scale, Tahoe-100M maps how various drug molecules impact different pathways, including immune-related pathways in diverse contexts (aka cancer models). We systematically explored an extensive array of these immune-related pathways to uncover drugs capable of modulating critical signaling mechanisms, including IFN-α and MHC-I. We designed a computational pipeline to evaluate hundreds of compounds simultaneously, allowing us to rapidly prioritize compounds based on their impact on these key immune pathways (Figure 2).
Figure 2. Computational pipeline illustrating method of prioritization of compounds for a given pathway of interest.
We found quite a few compounds that can effectively upregulate MHC-I. Some of these compounds were previously known immunomodulators (such as MEKi inhibitors). This provides valuable validation. But more importantly, it identified numerous new candidates (e.g., Elimusertib, an ATR kinase inhibitor, and TAK-901 , an Aurora kinase inhibitor) with previously unknown potential to stimulate immune recognition via MHC-I upregulation (Figures 3 and 4).
Figure 3. Tahoe-100M data showing compounds, grouped by MOA class, ranked by their effect on MHC-I geneset score upregulation.
Figure 4. Scatterplot of compounds by their perturbation effect on IFN-a and MHC-I geneset scores. Compounds are colored based on their MOA.
To get to this point, we only leveraged the data that is already in Tahoe-100M and the analysis pipeline we built on top of it; the question is whether orthogonal experimental assays would validate our findings. In other words, do independent experiments confirm that the newly identified compounds have the ability to upregulate MHC-I? The answer is “Indeed, they do”!
When we treated cells from CFPAC-1 cancer cell line with some of the new compounds we had identified, we saw marked increases in total MHC-I protein expression, as confirmed by western blotting (Figure 5). Flow cytometry analyses further validated that most selected compounds significantly enhanced MHC-I surface expression, underscoring their therapeutic potential to make tumors more visible and vulnerable to the immune system (Figure 6).
Figure 5. Experimental validation of total protein expression change after compound treatment. Western blot of CFPAC-1 cells after drug treatment and blotting for MHC-I shows upregulation for all compounds after drug treatment.
Figure 6. Experimental validation of cell surface MHC-I levels after compound treatment. Flow cytometry of CFPAC-1 cells with fluorescently labeled anti-MHCI antibody after drug treatment shows upregulation for some compounds after drug treatment. Normalized to vehicle treated (DMSO) cells.
These findings illustrate the unique capacity of the Tahoe-100M dataset to systematically address targeted biological questions—such as identifying drugs that specifically enhance MHC-I expression—at an unprecedented scale.
We have been saying consistently that gigascale, perturbative, single-cell maps like Tahoe-100M are a new modality of data for driving discovery. This example is a demonstration of that. Tahoe-100M allows cancer biologists to rapidly query their favorite biological pathways. It facilitates the discovery of novel therapeutic strategies and in this case, opens a new path for the development of precision-driven combinations aimed at significantly improving responses to ICIs in cancer treatment.