Accelerating Single-Cell Deep Learning with scDataset and Tahoe-100M
We at Tahoe love developer tools that make it easier to build on Tahoe-100M. This is a guest post highlighting the work of D. D'Ascenzo and S. Cultrera di Montesano, to be presented in ICML '25.
As single-cell technologies scale, biology is entering a new era—one where the volume and complexity of data now rival those in fields like computer vision and natural language processing. Among the most powerful advances are large-scale interventional datasets, which systematically measure how cells respond to genetic or chemical perturbations. These datasets make it possible to move beyond static descriptions of cell state, enabling researchers to uncover the mechanisms driving cellular behavior and to build models that predict how cells will respond to new conditions.
Tahoe‑100M (for the data, see Hugging Face) is the largest interventional single-cell dataset to date, comprising over 100 million transcriptomes that capture responses to 1,100 chemical perturbations across 50 cancer cell lines. The scale and richness of Tahoe-100M have already enabled new discoveries—for example, recent work used the dataset to identify compounds that enhance tumor visibility by upregulating MHC-I expression, a result that would have required extensive experimental screening in the past.
With datasets of this scale, AI and deep learning offer new possibilities for experimental biology. In the near term, machine learning models trained on interventional single-cell data can help researchers prioritize experiments, predict cellular responses to untested conditions, and reveal subtle patterns that might otherwise go unnoticed. Looking further ahead, these approaches hold promise for advancing our understanding of complex biological systems and for guiding the development of novel therapeutic strategies.
Why We Built scDataset
Despite the promise of large-scale interventional data, the practical reality of working with these datasets has been shaped by data storage and access constraints. The standard format for storing and sharing single-cell data is AnnData, which allows rich annotation and efficient storage of large, sparse matrices. However, most analysis tools and workflows for AnnData were designed for interactive exploration or modestly sized datasets—not for training deep learning models directly on tens or hundreds of millions of cells.
Traditional approaches often require loading the entire dataset into memory or converting to dense formats, quickly becoming infeasible at Tahoe-100M scale. As a result, the step from data collection to large-scale model training has remained a major bottleneck.
In our recent paper, we introduce scDataset, a PyTorch IterableDataset designed for efficient, scalable training on large single-cell omics datasets stored in the AnnData format. Unlike existing solutions, scDataset enables fast, memory-efficient, and shuffled data loading directly from disk, making it possible to train deep learning models on datasets as large as Tahoe-100M using standard hardware.
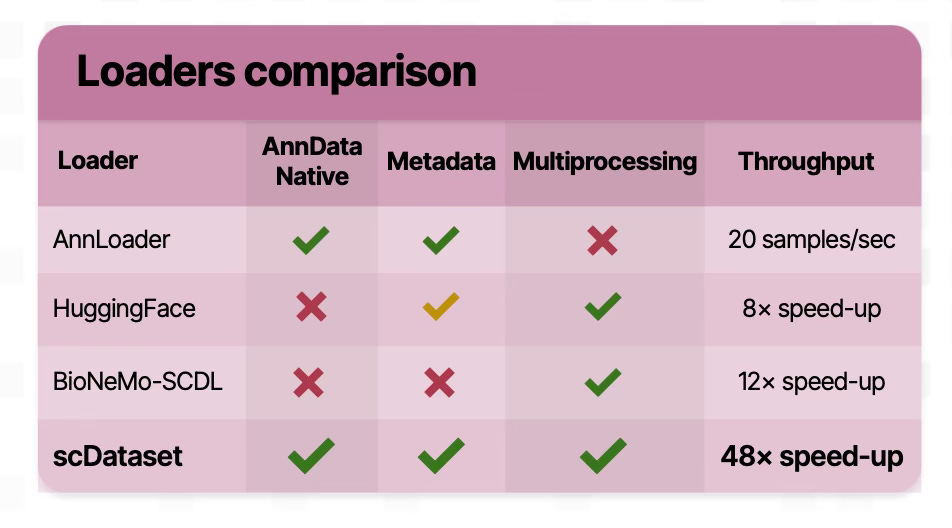
scDataset solves the AnnData bottleneck by combining block sampling and batched fetching, balancing randomness and I/O efficiency. In our benchmarks on the Tahoe-100M dataset, scDataset achieved up to a 48× speed-up over AnnLoader and substantial improvements over other popular data loading solutions (See below for a comparison with other loaders).
Getting Started with scDataset
To help the community get up and running, we've prepared a quickstart notebook that demonstrates how to use scDataset to efficiently load the Tahoe-100M dataset and train a simple linear classifier in PyTorch.
The notebook walks through:
- Loading Tahoe-100M .h5ad files
- Performing a stratified train/test split
- Wrapping the data with scDataset for efficient loading
- Training and evaluating a linear classifier for cell line recognition
You'll also find installation instructions, API documentation, and additional examples in the scDataset GitHub repository. If you have questions or feedback, feel free to reach out or open an issue on GitHub.
The Virtual Cell Challenge
The Arc Virtual Cell Challenge is the perfect venue to test the speed and scalability of scDataset in real-world applications. This open competition challenges the community to build predictive models of cellular responses using massive single-cell datasets. With scDataset, participants can focus on developing innovative models and analyses, confident that data loading will not be a limiting factor.
Whether you're new to large-scale single-cell analysis or aiming to accelerate your deep learning workflows, scDataset provides a practical and flexible foundation. We look forward to seeing what the community builds with scDataset and Tahoe-100M!